1. Introduction
Variant Call Format (VCF) files are a standard type of file used to represent information about variations within a genome. As with just about anything genomic, these files can get very large very quickly (e.g., dozens of gigabytes compressed), and the process of parsing them and converting them into any particular format can be conceptually simple yet very data-intensive.
This example illustrates the following points that were missing in the simple wordcount example:
- it uses a Python library specifically designed to parse VCF files that must be installed
- this Python library must be installed by a non-root user using a non-default version of Python
- this Python library is not designed to be parallel in any way
This guide builds upon my guide to the basics of using Hadoop streaming, so I recommend against trying to understand this guide without first going through my simple wordcount example. As with that guides, this guide comes with supporting material on GitHub which is meant to work on SDSC Gordon. I've also used semi-persistent Hadoop clusters on FutureGrid for much of the testing and development here, and this process should work on a wide range of clusters with minimal modification.
2. Conceptual Overview
Files don't always come in a format that is a perfect for map/reduce, and the output of your analysis doesn't always take the form of key/value pairs. Thus, I propose a few extra steps in the map/reduce pipeline highlighted in red:
- Proprocessing of data
- Mapping
- Shuffling
- Reducing
- Postprocessing of data
Within the context of the VCF files, preprocessing is necessary because the VCF file contains a header which describes the format of the rest of the file. Our mappers (or reducers) will all need to have this header information in order for our VCF reader library to make sense of the actual data that gets fed to it by Hadoop. Thus, the preprocessing step here involves reading the top of the input file and printing the header lines to stdout until the first non-header line is reached:
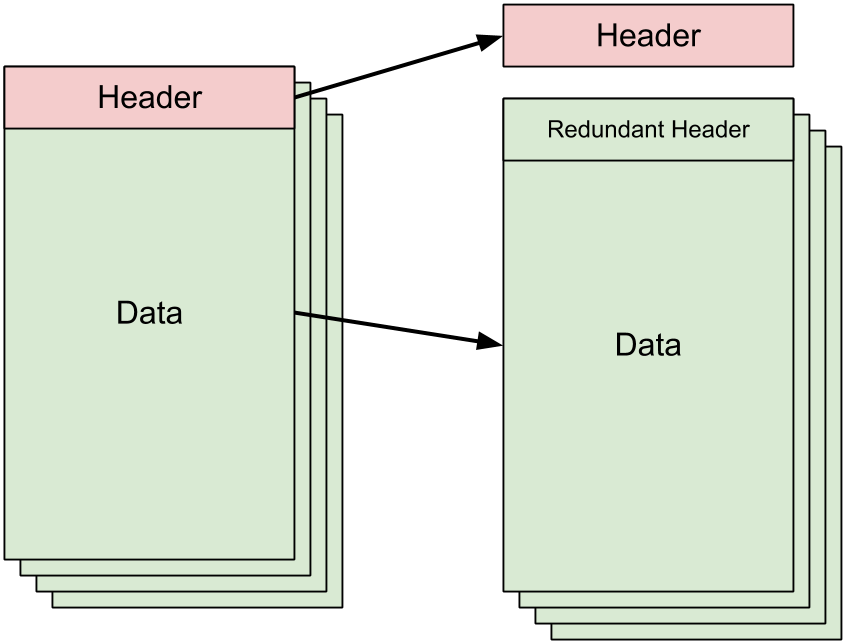
We copy this printed header information to a separate file that exists
outside of HDFS and don't bother messing with the actual VCF file's contents.
This obviates the need to actually read the entire VCF file since the act of
reading (or writing) that entire file is too big to do outside of HDFS. Thus,
the proprocessing step leaves us with a header file (header.txt)
and the original, untouched VCF input file.
The postprocessing step is dependent upon what we want to do with our VCF
data; when I was first getting into this application area, we were actually
using this process to transform the VCF data into a structured file that could
be COPY FROMed to a PostgreSQL database. This process of taking
the output from HDFS, connecting to the Postgres database, and COPY
FROMing that data was handled in the postprocessing step.
3. Install Python Library as Non-Root
Python makes it quite easy to install libraries within having root privileges on a machine. I recommend using VirtualEnv to this end; it essentially gives you a python installation that behaves as if you have administrative privileges, and it sets up a very easy one-line script to enable (or disable) its use.
The first step to setting up virtualenv is, of course, to download and unpack it:
$ wget http://pypi.python.org/packages/source/v/virtualenv/virtualenv-1.7.1.2.tar.gz $ tar xzf virtualenv-1.7.1.2.tar.gz
You then have to decide the prefix for your personal python installation. I
will pick ~/mypython, but this is arbitrary. You may decide you
want to have a separate Python virtualenv for each project. Either way, execute
the virtualenv.py script you just unpacked and tell it where you
want to set up camp. However, don't forget to load the
python/2.7.3 module first if you want this virtualenv to use
python2.7 instead of the system-default python2.4!
$ module load python/2.7.3 $ python virtualenv-1.7.1.2/virtualenv.py ~/mypython
This will create a bunch of files including
$HOME/mypython/bin$HOME/mypython/bin/python- your personal python executable that knows where to find the libraries you will be installing$HOME/mypython/bin/pip- the application you will use to install Python libraries$HOME/mypython/bin/activate- the script you'll source to activate this virtual environment$HOME/mypython/lib - where your python libraries will be installed
Then you can "enable" this virutal environment by sourcing the
activate script. The next time you log in and want to use this
virtual environment though, remember to module load python/2.7.3
first!
$ source ~/mypython/bin/activate
Your prompt will change to indicate to indicate that you're operating with
the Python virtualenv active. Then install all the Python libraries you want
using the pip command. This tutorial will require the pyvcf
library:
(mypython)$ pip install pyvcf Downloading/unpacking pyvcf Downloading PyVCF-0.6.4.tar.gz Running setup.py egg_info for package pyvcf ... Successfully installed pyvcf Cleaning up...
Now you can confirm that the library is accessible from within this virtualenv'ed python:
(mypython)$ python Python 2.7.3 (default, Feb 7 2013, 21:11:53) [GCC 4.1.2 20080704 (Red Hat 4.1.2-50)] on linux2 Type "help", "copyright", "credits" or "license" for more information. >>>import vcf >>>
4. The Mapper
This section is not done yet. You can see the entire mapper+reducer code in my github.
5. The Reducer
This section is not done yet. You can see the entire mapper+reducer code in my github.
6. Job Launch
There are a number of additions that must be made to our previous wordcount example to run this VCF parsing map job. Changes are highlighted in red below:
$ module load python/2.7.3 $ $HOME/mypython/bin/python $PWD/parsevcf.py -b patientData.vcf > header.txt $ hadoop dfs -mkdir data $ hadoop dfs -copyFromLocal ./patientData.vcf data/ $ hadoop \ jar /opt/hadoop/contrib/streaming/hadoop-streaming-1.0.3.jar \ -D mapred.map.tasks=4 \ -D mapred.reduce.tasks=0 \ -mapper "$HOME/mypython/bin/python $PWD/parsevcf.py -m header.txt,0.30" \ -reducer ">python $PWD/parsevcf.py -r" \ -input "data/patientData.vcf" \ -output "data/output" \ -cmdenv LD_LIBRARY_PATH=$LD_LIBRARY_PATH
The changes highlighted do the following:
- module load python/2.7.3 is necessary since our vcf library was installed for the python version provided by this module (2.7.x), not the version provided by the system (2.4.x)
- python $PWD/parsevcf.py -b ... is the preprocessing step we added to extract the VCF header into a separate file
- -D mapred.map.tasks=4 is really optional; it tells Hadoop that we would like (but won't necessarily get) four mappers
- -D mapred.reduce.tasks=0 is what disables the shuffle and reduce steps, since our example doesn't need to reduce anything
- We explicitly specify the path to our custom Python ($HOME/mypython/bin/python ...) when
calling our mapper/reducer scripts to ensure that Hadoop uses our custom
Python that has the PyVCF library installed. The
-m header.txt,0.30 is passed because our mapper function expects
a comma-separated list containing
- the file containing just the VCF's header section (
header.txt) - the allele frequency above which a record gets printed (
0.30)
- the file containing just the VCF's header section (
- We don't need to specify a -reducer since we set
mapred.reduce.tasks=0 - -cmdenv LD_LIBRARY_PATH=$LD_LIBRARY_PATH is critical
and passes the contents of our environment's $LD_LIBRARY_PATH
variable to the Hadoop streaming execution environment. We modified the
contents of this $LD_LIBRARY_PATH when we issued module load
python/2.7.3, so this
-cmdenvoption ensures that the changes made when we loaded that module are propagated out to Hadoop. You can specify multiple-cmdenvoptions if you need to propagate other environment variables that your mappers/reducers may need.
7. Scaling Behavior
This section is not done yet.
This document was developed with support from the National Science Foundation (NSF) under Grant No. 0910812 to Indiana University for "FutureGrid: An Experimental, High-Performance Grid Test-bed." Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the NSF.