4. Introduction
The foreach package, which is authored by the folks at Revolution Analytics, is functionally equivalent to the lapply-based parallelism discussed in the previous section, but it exposes the parallel functionality to you (the programmer) in what may seem like a more intuitive and uniform way. In particular,
- you do not have to evaluate a function on each input object; rather, the code
contained in the body of the
foreachloop is what gets executed on each object - your parallel code uses the same syntax across all of the possible parallel
backends, so there is no need to switch between
lapply,mclapply, andparLapply
Perhaps it is more sensible to illustrate what this means by comparing the
equivalent commands expressed using lapply and
foreach:
mylist <- c( 1, 2, 3, 4, 5 ) output1 <- lapply( mylist, FUN=function(x) { y = x + 1; y } ) output2 <- foreach(x = mlist) %do% { y = x + 1; y }
Both codes iterate through every element in mylist and return a list of the same length as mylist containing each of the elements in mylist plus one.
However, there is an implicit assumption that there are no side effects of the code being executed within the foreach loop--that is, we assume that the contents of the loop body are not changing any variables or somehow affecting any global state of the rest of our code. For example, the variable y is not guaranteed to contain 6 at the end of the foreach loop, and in fact, it is not even guaranteed to be defined.
4.1. Halfway to parallel
Going back to the simple k-means example, recall that this is what the most simplified (serial) version of the code would look like:
data <- read.csv('dataset.csv') result <- kmeans(data, centers=4, nstart=100) print(result)
As with lapply-based parallelism, we have to take a step backwards to make the code amenable to parallelization. The foreach version of the k-means code looks like this:
library(foreach) data <- read.csv('dataset.csv') results <- foreach( i = c(25,25,25,25) ) %do% { kmeans( x=data, centers=4, nstart=i ) } temp.vector <- sapply( results, function(result) { result$tot.withinss } ) result <- results[[which.min(temp.vector)]] print(result)
Again, this is significantly more clunky than the simple three-line serial k-means code; I've highlighted the differences in blue above. However, if we compare it to the lapply-style version of our k-means code, it is actually quite similar. Here is the same foreach code with the differences from lapply now highlighted:
library(foreach) data <- read.csv('dataset.csv') results <- foreach( i = c(25,25,25,25) ) %do% { kmeans( x=data, centers=4, nstart=i ) } temp.vector <- sapply( results, function(result) { result$tot.withinss } ) result <- results[[which.min(temp.vector)]] print(result)
4.2. doMC: shared-memory parallelism
Once we've coerced our algorithm into the foreach-based formulation, it
becomes very easy to parallelize it. Instead of making a new call
parallelized form of lapply like mclapply, foreach
lets us register a parallel backend by loading the appropriate backend
library and registering it:
library(foreach) library(doMC) data <- read.csv('dataset.csv') registerDoMC(4) results <- foreach( i = c(25,25,25,25) ) %dopar% { kmeans( x=data, centers=4, nstart=i ) } temp.vector <- sapply( results, function(result) { result$tot.withinss } ) result <- results[[which.min(temp.vector)]] print(result)
The doMC library is what provides the "multicore" parallel
backend for the foreach library. Once loaded, all you have to do
to parallelize your loop is call registerDoMC to indicate the
number of cores to use (four in the above example) and replace the %do
with %dopar% to tell foreach to use the parallel
backend you just registered.
As one would hope, using foreach with the doMC
parallel backend provides the same speedup as mclapply:
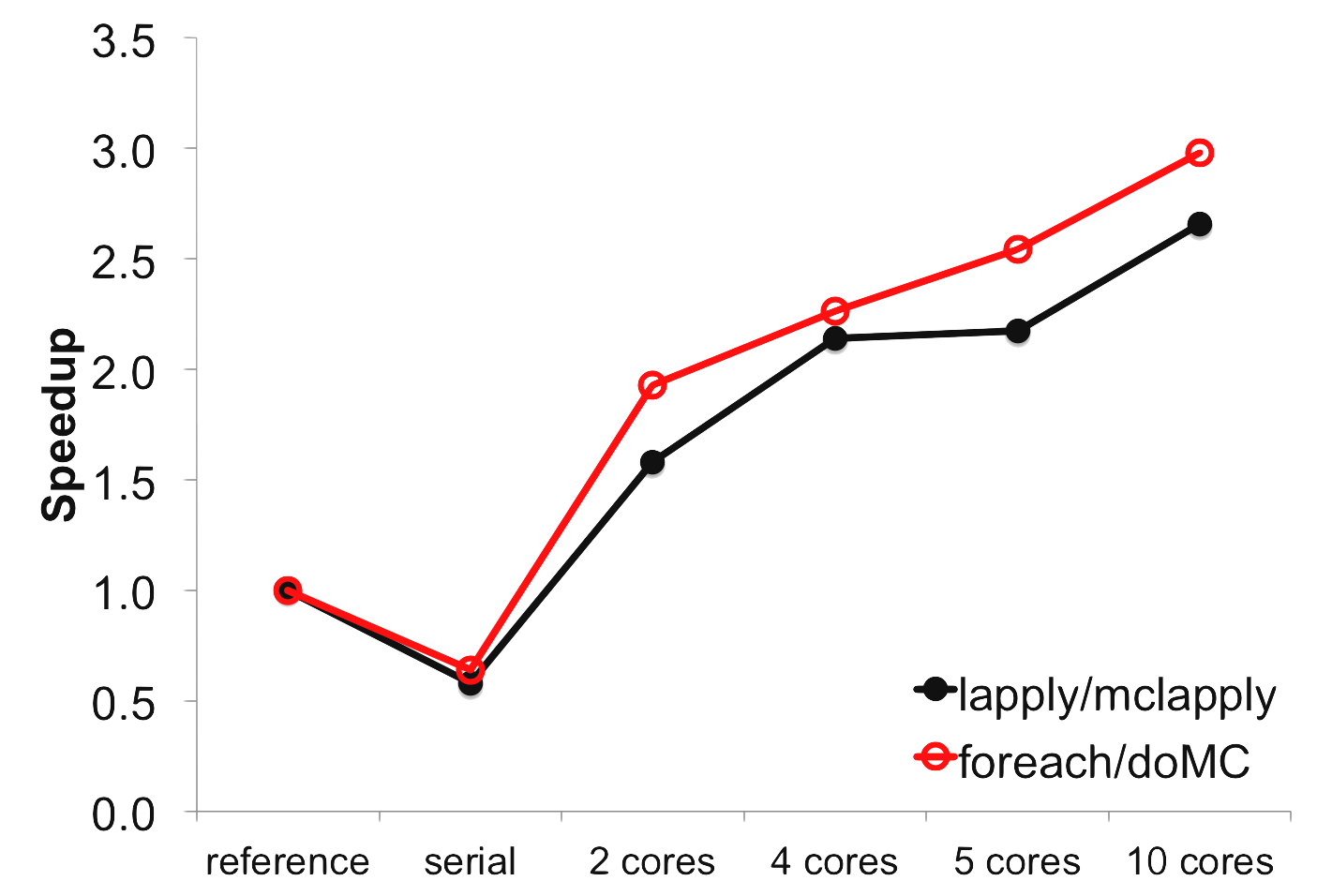
The slightly greater speedup in the foreach case (red line) is not significant since the dataset I used
is a bit of a trivial case and only took a few seconds to run.
4.3. doSNOW: distributed-memory parallelism
Just as we used the snow library to perform multi-node,
distributed-memory parallelization with parLapply, we can use the
doSNOW parallel backend with foreach to perform
distributed-memory parallelization. Here is what our k-means example would look
like:
library(foreach) library(doSNOW) data <- read.csv('dataset.csv') cl <- makeCluster( mpi.universe.size(), type='MPI' ) clusterExport(cl,c('data')) registerDoSNOW(cl) results <- foreach( i = c(25,25,25,25) ) %dopar% { kmeans( x=data, centers=4, nstart=i ) } temp.vector <- sapply( results, function(result) { result$tot.withinss } ) result <- results[[which.min(temp.vector)]] print(result) stopCluster(cl) mpi.exit()
The differences between the doMC and doSNOW version
of this foreach k-means example are highlighted in blue. We really only had to
make three changes:
- we replaced
doMCwithdoSNOWand used the corresponding backend registration function,registerDoSNOW - we had to create a cluster object just like we did with
parLapply - we also had to export our input data to all worker nodes using
clusterExport, also like we did withparLapply
Everything we learned about the types of clusters we can create for
parLapply also work with the doSNOW backend for
foreach. It follows that we also have to be mindful of distributing all of
the data we will need our workers to see via the clusterExport
call when using MPI or SOCK clusters. At the end of the day though, there
aren't any fundamental differences between the things we can do with these
cluster objects when moving from parLapply to doSNOW.
5. Caveats with lapply- and foreach-based parallelism
Ultimately, using these lapply- and foreach-style approaches to parallelizing your R scripts works well if your R script is very CPU-intensive and spends a lot of time just doing processing on a relatively small-ish dataset. I often see people use this approach under conditions when they want to
- try many different statistical models on the same set of data
- run the same statistical model on many different datasets
- ...or just about any other case where many independent calculations must be performed
The biggest downside to using these methods is that you are still limited by the amount of memory you have in a single node as far as how large of a dataset you can process. This is because you still load the entire dataset into memory on the master process:
data <- read.csv( 'dataset.csv' )
and clone it across all of your nodes:
clusterExport(cl, c('data'))
Thus, these approaches will not really help you solve problems that are too large to fit into memory; in such cases, you either need to turn to special out-of-core packages (e.g., the ff or bigmemory libraries) or turn to map/reduce-style parallelism with a framework like Hadoop or Spark. There are programming tricks you can play to skirt around this (e.g., loading subsets of the input data, distributing it, deleting that object, and then garbage collecting before loading the next subset) but they rapidly become extremely complicated.
These approaches to parallelism so far require explicitly calling specific parallel libraries and deliberately restructuring algorithms to make them amenable to parallelization. In the next section, we will examine alternative forms of parallelism that do not require such significant changes to code.